UCSC Treehouse
Public Data
The Treehouse Childhood Cancer Initiative is a research arm of the UCSC Genomics Institute. We enable the sharing of pediatric cancer genomic data using tools developed by our Genomics Institute colleagues. We use shared data to analyze a child’s tumor against both child and adult patient cancer tumors using a “pan cancer” or cross-comparison gene expression analysis. Our goal is to identify situations where an approved drug, often an adult drug, is predicted to work on a child with cancer.
As part of our research, we have gathered a compendium of RNA gene expression data which we have made available for download and visualization.
Our samples are derived from partner clinical sites and publicly available repositories, including TARGET and TCGA. Expression data from over 11,000 samples is available along with clinical data including age, gender, and disease type.

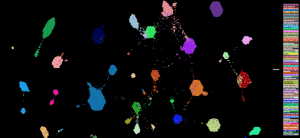
Tumormap
The UCSC TumorMap interactively displays samples in the Treehouse dataset positioned according to their RNA profiles. Users can color the samples based on dataset features like Disease. This browser shows samples clustered using the OpenOrd algorithm and best separates smaller groups. (See“TumorMap: Exploring the Molecular Similarities of Cancer Samples in an Interactive Portal.” Cancer Research November 2017).
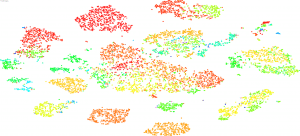
Cluster Browser
The UCSC Cluster Browser interactively displays samples in the Treehouse dataset positioned according to their RNA profiles. Users can color the samples based on dataset features like Disease. This browser quickly shows samples clustered using the t-SNE algorithm and best shows relationships among larger groups.
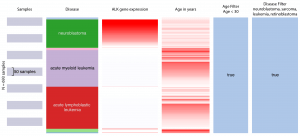
Xena
UCSC Xena allows users to explore the Treehouse dataset, finding correlations and trends within and across genomic and phenotypic variables. Users can interactively add, remove, and rearrange arbitrary slices of data including genes, transcripts and other dataset features. This example from our July 2017 dataset shows that neuroblastoma in comparison to other pediatric cancers has a much stronger ALK gene expression and younger patient population.
Files
Limited Clinical Data
Age, gender, and disease are provided for RNASeq samples compiled by the UCSC Treehouse Childhood Cancer Initiative. Samples derived from clinical sites, publicly available repositories, TARGET, and TCGA.
TPM Gene Expression, log2-Normalized
Values in this dataset use HUGO gene names and are TPM, transformed by log2(x+1) of the TPM value.
Expected Counts Gene Expression
Values in this dataset are expected_count and use Ensembl gene IDs.
Download
Compendium v5 Public (April 2018)
Visualize
- Tumormap
- Cluster Browser (coming soon)
- Xena
Files
- Clinical Data
- TPM Expression
- Expected Counts Expression (coming soon)
This compendium was released in April 2018. It includes a total of 11,258 samples from Treehouse (identifiers start with “TH”), TCGA and TARGET projects. This data was generated by library preparation methods including polyA selection and ribosomal depletion.
Compendium v4 Public (July 2017)
Visualize
- Tumormap
- Cluster Browser (coming soon)
- Xena
Files
- Clinical Data
- TPM Expression
- Expected Counts Expression (coming soon)
This compendium was released in April 2018. It includes a total of 11,258 samples from Treehouse (identifiers start with “TH”), TCGA and TARGET projects. This data was generated by library preparation methods including polyA selection and ribosomal depletion.
Pipeline
We are committed to data sharing and encourage you to be part of this sharing network. The data provided here was processed with the RNA-Seq pipeline developed by the UC Santa Cruz Computational Genomics Lab. The pipeline is available for general use; the source code is hosted on GitHub at BD2KGenomics/toil-rnaseq and a Dockerized version is available at UCSC-Treehouse/pipelines.
Contribute
If you use our data, and have data of your own, pay it forward by running our pipeline and sharing back. We will add these results to our public compendium of expression data, with a credit to your contribution. By doing so, your samples and those of other partner sites will contribute to an ever-improving virtuous cycle of data sharing, ensuring that each participant’s data pays it forward to future participants!
Acknowledgements
We are grateful to all our supporters and clinical partners (see below, and on our Acknowledgments page). Without them, we would not be able to accomplish this important work.
Thank you to all who are sharing data. A special shout out to the St. Baldrick’s Foundation and the California Initiative to Advance Precision Medicine, not only for supporting Treehouse but for their commitment to data sharing and their efforts to advance responsible data sharing.








Data Usage Policy
If you use our pipeline to process your data, we would appreciate it if you share the results with us, so it can be added to the public database. Just send us an email and we’ll get in touch to arrange the data transfer. Our goal is to benefit researchers and pediatric patients everywhere through access to data.
